Author: Richard Challis
-
Introducing BlobToolKit
BlobToolKit is a set of tools to allow interactive exploration, assessment and filtering of genomic datasets. The main tools are BlobTools2, a re-implementation of BobTools to support additional features and The BlobToolKit Viewer, which allows interactive exploration of datasets. This series of blog posts aims to introduce many of the features in BlobToolKit and provide…
-
Genome Science 2018
We will be presenting a poster describing BlobToolKit at Genome Science 2018 For more details, either browse this site or take a look at the project code on GitHub.
-
BlobToolKit Viewer
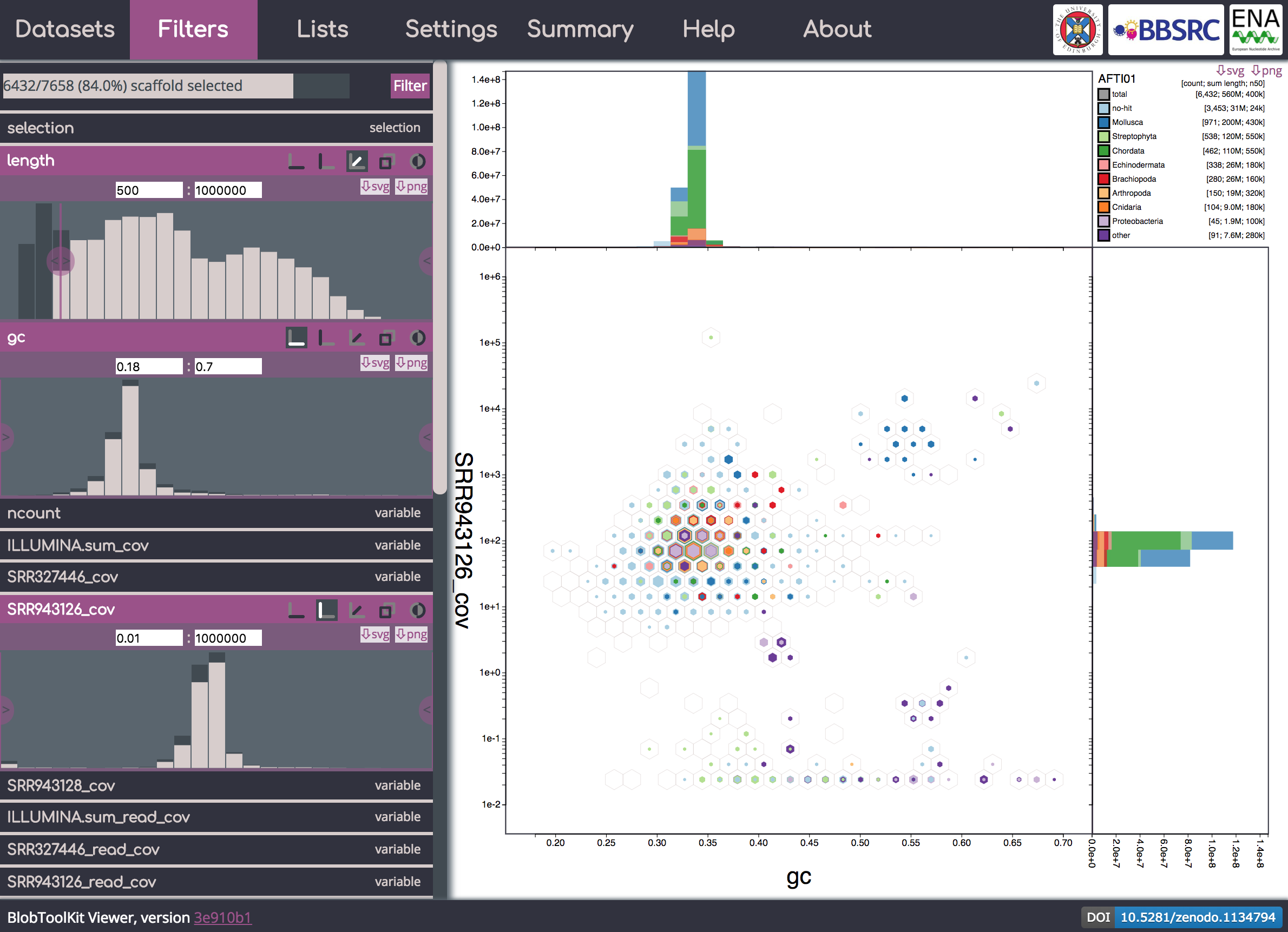
Visualise assemblies using the new BlobToolKit viewer (blobtoolkit.genomehubs.org/view/, github.com/blobtoolkit/viewer): Example using data obtained from ENA for the Pacific oyster Crassostrea gigas (AFTI01). Processed using a BlobToolKit Snakemake pipeline available at github.com/blobtoolkit/insdc-pipeline.
-
BlobTools manuscript now available
Dom Laetsch and Mark Blaxter’s BlobTools manuscript1 is now available on F1000 Research. This Manuscript describes BlobTools version 1, which is available at github.com/DRL/blobtools. 1 Laetsch & Blaxter 2017. BlobTools: Interrogation of genome assemblies [version 1; referees: awaiting peer review]. F1000Research, 6:1287 (doi: 10.12688/f1000research.12232.1)
-

Introducing BlobToolKit
The BlobToolKit project has been funded by the BBSRC for 3 years to develop the latest iteration of the Blobology/Blobtools genome assembly QC, visualisation and filtering tools. The project is a collaboration between the research groups of Mark Blaxter at the University of Edinburgh and Guy Cochrane at EMBL-EBI to develop software tools and protocols…